D-Lib Magazine
January/February 2017
Volume 23, Number 1/2
Table of Contents
Conquaire: Towards an Architecture Supporting Continuous Quality Control to Ensure Reproducibility of Research
Vidya Ayer
CITEC, Bielefeld University
http://orcid.org/0000-0002-3661-3277
vayer@techfak.uni-bielefeld.de
Christian Pietsch
Bielefeld University Library
http://orcid.org/0000-0001-8778-1273
cpietsch@uni-bielefeld.de
Johanna Vompras
Bielefeld University Library
http://orcid.org/0000-0002-8224-2884
johanna.vompras@uni-bielefeld.de
Jochen Schirrwagen
Bielefeld University Library
http://orcid.org/0000-0002-0458-1004
jochen.schirrwagen@uni-bielefeld.de
Cord Wiljes
CITEC, Bielefeld University
http://orcid.org/0000-0003-2528-5391
cwiljes@techfak.uni-bielefeld.de
Najko Jahn
Bielefeld University Library
najko.jahn@uni-bielefeld.de
Philipp Cimiano
CITEC, Bielefeld University
cimiano@techfak.uni-bielefeld.de
Corresponding Author: Vidya Ayer, vayer@techfak.uni-bielefeld.de
https://doi.org/10.1045/january2017-ayer
Abstract
Analytical reproducibility in scientific research has become a keenly discussed topic within scientific research organizations and acknowledged as an important and fundamental goal to strive for. Recently published scientific studies have found that irreproducibility is widely prevalent within the research community, even after releasing data openly. At Bielefeld University, nine research project groups from varied disciplines have embarked on a "reproducibility" journey by collaborating on the Conquaire project as case study partners. This paper introduces the Conquaire project. In particular, we describe the goals and objectives of the project as well as the underlying system architecture which relies on a DCVS system for storing data, and on continuous integration principles to foster data quality. We describe a first prototype implementation of the system and discuss a running example which illustrates the functionality and behaviour of the system.
Keywords: Conquaire, Analytical Reproducibility, Quality Control, Reproducible Computational Research, DVCS, Computational Science, Research Data Management System, Data Science, Infrastructure Architecture
1 Introduction
Reproducibility of scientific research is an essential principle within science. Scientific results should stand the scrutiny of the research community and should be verifiable by peers. In practice, however, reproducing or confirming previous research results can be a major challenge. In fact, meta-studies in psychology, medicine and also computer science have documented the challenges involved therein and they have empirically shown that the success rate in reproduction of research results is low.
A low success rate in reproducibility has been particularly observed in psychology. A recent study has shown that only 39% of published studies can be regarded as being reproducible [1]. In pharmaceutical research, success rates are even lower at around 18% for clinical trials [2] second phase. Nature's most recent survey [3] carried out among 1,576 scientists has revealed that in more than 70% of the cases researchers fail to reproduce the experiments of other scientists. Furthermore, they are unable to reproduce their own experiments in around half of the cases.
Failure to reproduce scientific results can have many causes. For instance, the differences can be due to the experimental setup, the measurement methods used, the scientific equipment, the calibration of scientific instrumentation or simply due to different samples of the population under study which are seldom directly comparable.
An important step in the generation of scientific results lies in the computational analysis of the primary data or derived secondary data. In most cases, software packages (such as SPSS, R, Excel, SAS, Stata) are used in this part of the process to test a hypotheses by performing some computational or statistical analysis on the derived primary data. We will refer to this part of the process as the "analytical phase". Complete reproducibility of an experiment can be extremely difficult. However, reproducing the "analytical phase" seems less difficult, as it would essentially require access to the primary and/or derived data and to the analytical tools used by the researchers to derive some result.
Thus, it appears feasible to strive for a more reduced version of reproducibility that we will refer to as "analytical reproducibility" in order to ensure that a third party researcher could reproduce the computational/statistical analysis performed on derived data to yield a particular conclusion, thereby being able to independently verify the results and research hypothesis. This would be a significant step, in our view, towards supporting reproducibility in research. However, the mere availability of data is not enough to ensure analytical reproducibility. In order to be able to reproduce an analysis, the data needs to be of high quality, in the sense that it does not contain syntactic or semantic errors, is well documented, and the script or code that was used to analyze the data is also openly available.
2 Conquaire Project Objectives
The core objective of the Conquaire (Continuous Quality control for research data to ensure Reproducibility) project [4] is to support research reproducibility by extending the technical infrastructure available at Bielefeld University with the underlying goal of creating a generalized research data management system (RDMS) to manage the data and scripts for publications that other research institutions can adopt. In order to support researchers in meeting the open data and open research standards, the goal of the Conquaire project is to develop a generic infrastructure framework that can support scientists towards achieving analytical reproducibility. One of the project goals is to enumerate the steps researchers can take in order to ensure that their scientific data, the scripts and the associated computational analyses, can be of value long after the research project has concluded. Over the past few years, there have been numerous publications on the challenges in the field of data sharing, open data, analytical research, reproducibility, privacy, etc.
As described above, within the Conquaire project we are concerned with developing an institutional infrastructure that supports reaching "analytical reproducibility". At the heart of the Conquaire project is the requirement of ensuring that researchers deposit their data, code, etc., in a version control system that supports tracking versions of code and data and labelling snapshots with persistent identifiers. Snapshots in this sense are important versions, such as the one submitted to a journal for reviewing. However, storage and versioning are not enough. In order to ensure reproducibility by others, the project desiderate several other functionalities:
- Researchers need to make their research data available in some common format so that others can download and inspect the data.
- Data needs to be syntactically valid according to common open format standards.
- Data needs to be sufficiently documented so that third parties can understand the exact conditions under which the experiments were carried out and each data element being documented for users to understand its semantics.
- The analytical processes for data processing need to be made available together with the research data.
- An independent researcher or analyst needs to be able to rerun the analytical process to independently verify the results reported in the paper.
- The computational software tools used by the researcher must be free/libre software that is publicly available for everyone to use.
The crucial aspect of the above-mentioned points are monitoring data quality (e.g. syntactic validity), adherence to best practices of the community, passing semantic integrity tests, etc. Our goal in the Conquaire project is to foster an agile approach to research data management in which data quality is continuously monitored and ensured right from the start of the research cycle. So far, in most scientific projects, publishing data and making results available is typically only an afterthought and are typically done only when the research is actually finished and results have been published in a written article. In general, once the research project has concluded, there is understandably very little motivation for researchers to invest in subsequent data management to make their analysis and results reproducible by others.
Therefore, in Conquaire, we intend to move data publication to the heart of the research process, creating incentives for a researcher to make their papers "executable" and "reproducible" right from the start of their research process. Our objective is to support researchers with the appropriate workflows and incentive mechanisms that support them in this task. We envision the following: researchers create a version control system repository for their research project and build their research work on our envisioned infrastructure, which will be simplified to allow users to perform tasks via GUIs, shielding researchers from the technical aspects. By creating a research project, a basic folder structure will be created consisting of (i) documentation, (ii) data and (iii) analysis. These folders will correspondingly contain the main elements needed for analytical reproducibility. At all stages, the researchers will get a visual representation of the status of the project in terms of achieving the goal of full analytical reproducibility, with indication of actions needed to be performed in order to achieve the status of full analytical reproducibility, including indications of which quality test the data has passed.
Full reproducibility will be only achieved if an independent third party confirms that the results of the analytical processes run on the data provided actually return the results as described in a given paper. Beyond the technical hosting support we provide, there are clear incentives for researchers to take part in this project. One important incentive for a researcher during the project will be the support for ensuring data quality in the workflow, and ensuring that the data follows certain constraints from the very beginning of their research project.
A further incentive for case study partners participating in the Conquaire project is the fact that they are deeply convinced about analytical reproducibility, want to learn best practices that can help them improve their research workflow and want to manage their research shared with third parties or interdisciplinary groups within their fields. This is a deeper incentive system that will encourage them to collaborate as responsible researchers more effectively than using superficial incentives, such as, a reward system of badges, ranking, upvoting, or a points-based system for certain tasks or milestones achieved as part of the gamification concept. Finally, for a researcher, third party verification and validation (supported through a version control system (VCS)) will help to ensure that there are no hidden or unwarranted assumptions, nor flaws in data collection, analysis, etc. This will contribute to the higher quality research results that researchers strive for.
In this paper, we briefly describe the first version of the Conquaire architecture and the type of quality checks and standards it supports by default. We then present a simple proof-of-concept implementation that illustrates the process. We conclude by highlighting future developments and briefly describe our collaboration with the research projects that we intend to support within the project as case study partners.
3 Case Studies
The central idea of the Conquaire project is the close co-operation with nine partner projects who serve as case studies to define requirements and constantly test the system during the software development cycle. The following table showcases the interdisciplinary scientific research fields vis-a vis the software, data formats and other project tools used by the project partners:
Table 1: Software tools and data formats used by the project partners.
| Partner discipline |
Biology, Computer Science, Applied Computational Linguistics, Neurobiology, Sports Science, Neurocognitive Psychology, Atmospheric & Physical Chemistry, Economics and Linguistics. |
| Software |
Python (Pandas), R-lang, C, C++, Matlab, SPSS |
| VCS |
Git, SVN |
| Database |
MySQL |
| Data Formats |
CSV, XLS, XML, JSON, JPEG, MP4, EAF (ELAN annotated files) |
| Supercomputing |
HPC cluster |
| Data Storage |
Dropbox, Sciebo, private servers, backup drives, etc. |
Our discussions with the partners provide us with insights into the challenges they face with respect to data exchange between collaborators. Like-minded research projects that are collaborating on the same research problem would like to share some data among themselves, especially their preliminary research observations or analysed output files. Currently, this is a challenge as there is no single infrastructure that helps them accomplish this, resulting in ad hoc strategies that are neither consistent nor particularly systematic.
For example, the researcher may use Dropbox to share data between the collaborating Universities, but this approach only allows them to dump a large batch of files online which does not give a researcher any important information about the shared data, such as:
- What values of analysed data has changed between two (or more) data dumps into Dropbox, and how that change affects their ongoing research.
- The ability to revert or experiment with data over the research project timeframe — e.g., after 5 data uploads to Dropbox, if a collaborator wants to access an older (e.g., second) analysis for comparing data points, or use the data within their current work, the task of data retrieval turns into a challenging time-sink as there is no method to track revisions of multiple uploads.
- The lack of timestamps and version control makes it an extremely challenging task to trace the timeline and purpose of any changes made to the data, results and program scripts, within the lifecycle of a research project.
Our case study partners have just commenced their research projects, hence their research data management plan (RDMP) will be an ongoing one. We have started requesting them for sample data files in order to understand their data needs. Thereafter, we plan to identify common technology patterns that can be abstracted for quality checks and other standards, as done by peer OpenData groups. With respect to publishing data openly, as explained in detail in the 'software architecture' section, we plan to integrate the Conquaire server with the existing publication repository PUB at Bielefeld University.
4 Conquaire Architecture
The architecture of the Conquaire system has been designed to support continuous quality control of research data. The abstract architecture, as depicted in Figure 1, assumes the researchers will commit their data and program scripts into a VCS. Besides providing a central, backed up storage for their data, the VCS comes with the advantage that data and software is versioned, so that a particular version of the data and script can be uniquely referenced. Once researchers deposit data into the VCS, the Conquaire server will analyze the data, and depending on the MIME type/extension of the data, apply some checks by invoking the Quality Control Framework. The Quality framework is essentially a middleware layer that calls different quality control processes for certain types of data files. As a proof-of-concept, we have so far implemented a quality control that performs a number of checks on CSV files (see below).
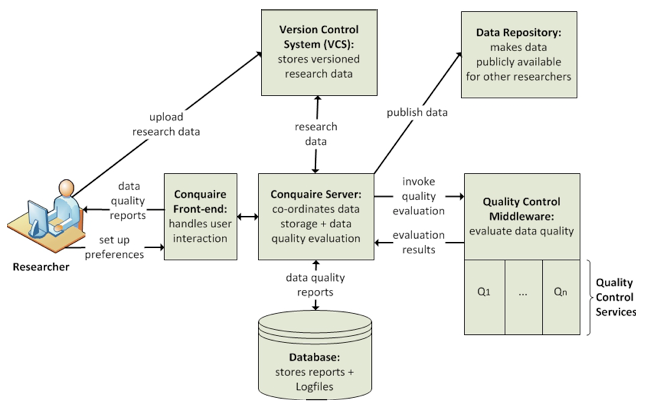
Figure 1: Conquaire system architecture
The main components of the Conquaire architecture are the following:
GUI/Frontend: The GUI is the visual interaction interface enabling the researcher to login, create new repositories, add data to the repository, etc. Here, they can read and inspect the status messages displayed, for example, the quality reports from the quality control middleware to the Conquaire server (see below) that continuously monitors their repository for data changes to improve the data "readiness" for reuse.
VCS Server: The VCS server supports versioned storage of the data by using a standard distributed version control protocol, such as Git or Mercurial, that implements standard VCS functionality such as:
- Multiple "central" repositories, with a large distributed development model that uses a reference copy of the codebase as default and stores only working copies.
- A non-linear development workflow that supports rapid branching and merging with every project collaborator having a local copy of the entire development history.
- Allowing all collaborators to keep their working changes separate as additional development branches within the same working directory.
- Each working copy effectively functions as a remote backup of the codebase and of its change-history, protecting against data loss.
- Local operations, such as, commits, merges, and reverting changes are fast, because there is no need to communicate with a central server.
- Cryptographic support for commit history ensures that a salted hash ID for a particular version (a commit in VCS terms) is stored for the tree.
Conquaire Server: This is the heart of the Conquaire architecture. It monitors different repositories and reacts to any change(s) in the repository, such as modified files being committed or new files being checked in. On the basis of the data file extension, it invokes the quality control middleware asking it to apply a certain set of checks to the given data file. Upon receiving the results from a specific quality process, the server displays the quality report (success/ error message) to the researcher via the GUI / Frontend.
Quality Control Middleware: This middleware component will be pre-programmed to be "aware" of different quality control standards, the registered checks and processes, and can pass data to a specific process to perform the appropriate quality check on the data and provide continuous feedback for each data commit or file(s) committed to the VCS.
Quality Control Services: These are a number of quality control processes registered at the quality control middleware to perform tests and checks on the data, then return the result to the server for storage.
Database: The database is used to store the results of all the quality control processes performed with a timestamp of their result. It also logs and stores the messages generated by the system with information about which bugs (issues) have been solved, closed, etc.
Trusted Data Repository: Bielefeld University's institutional repository PUB already allows researchers to upload data publications as well as traditional publications. Both kinds of publications are assigned a persistent Digital Object Identifier (DOI) on upload, and relationships between them are recorded and exposed in the DataCite [5] metadata format. Changes in bibliographic metadata are automatically tracked in the VCS. Researchers are encouraged to upload full texts and data sets to PUB, where they are stored safely: Nightly backups are stored on tape by the computing centre. Additionally, open access publications are preserved in a geographically distributed archival network called SAFE PLN, which runs the LOCKSS software. By the end of the Conquaire project (2019), we will also preserve other publications including research data in SAFE PLN.
5 Implementation
In our first implementation of the Conquaire architecture, the components will be implemented as follows:
Front end: We will have a friendly GUI for the researchers which will provide synchronized information about their VCS commits as well as the data quality information. For example, researchers with clean data will get a badge (star rating system) that will incentivize the production and storage of clean data.
VCS Server: The backend server running a distributed VCS stores data objects which will support the non-linear development workflow of branching and merging with every project collaborator. It will store the research data submitted by the nine case study partners and communicate with message-oriented middleware infrastructure to send and receive messages within the distributed systems that are triggered when any researcher(s) checks-in data or scripts into the VCS. When a researcher makes a commit, it will email the project members the information about the checked in files. This is tracked via the explicit object packing feature in a VCS, where each commit is an object, stored in a single separate file called 'pack-file' that is compressed, and a corresponding index file is created for each pack-file. The daemons and runners will continuously monitor and execute the server-side scheduling and logging process.
Conquaire Server: The server is the backend library framework that will play an active role in monitoring and controlling the data and resulting artifacts being committed into the VCS. The stateful part has logic to make process decisions and communicate with the registered members. Currently, we are in the development stage with a pre-alpha version that abstracts from the GUI, and when a researcher commits the data, it will email all the project members the commit information. Continuous integration runners monitoring the server for data commits will trigger the Quality library to perform checks and provide continuous feedback for all data changes.
Quality Control Middleware: The Quality monitoring server is a framework that is responsible for monitoring all the data checked in for data quality, data management, data curation, data analysis and processing. The Quality framework will monitor the project data quality for all the metadata in a pre-decided specific format that adheres to certain standards, e.g., ISO, IETF, W3C standards etc. This will allow us to filter the data for certain basic standards for different file formats explicitly and run some standard tests.
Conquaire Database: The Database for the Conquaire server will not store data files, rather it will only store metadata information and information data tags in the database in a semantic data format. The daemons and message-oriented middleware monitoring the VCS server would have triggered the Quality framework to act on the file, and once the checks are complete, the information would be recorded and stored. The quality check standards for the respective research data will be provided by each case-study partner. For example, a file with an extension (e.g., .csv, .xml, or .eaf) will have a corresponding script codified with the standard tests that will run against the data file with the metadata stored in a graph database.
6 First Quality Control Strategy & Next Steps
As a first proof-of-concept of our quality monitoring component we have implemented a tool that checks CSV files for compliance with certain best practices. The idea is that when a CSV file is committed to the VCS, the quality tests are automatically executed to ensure compliance with a number of good practices described in the "Good Enough Practices for Scientific Computing" [6] and Software Carpentry [7] papers. We assume that every CSV file is accompanied by a metadata schema file describing the data in the human-readable YAML format, such as "csvy". If this metadata file is not available, we automatically insert a template into the VCS server for the researcher to fill in.
In particular, we apply checks to ensure the following:
- Each column in the data has a name that appears in a dictionary (otherwise, a warning is issued to the researcher).
- The range of values is specified in the metadata file.
- A textual definition of the column is available (a warning is generated if the definition is less than 5 tokens).
- No value is out of range.
- Mathematical operations (min, max, average, standard deviation) on columns containing numerical data measurements.
- A data type for each value is inferred and all values match the datatype.
As a running example, we use the following CSV data describing Air Quality in New York, with the data descriptions. Assuming that an individual researcher commits the above CSV file into the data directory of a new VCS repository, they would receive a warning message alerting them that the data has no schema. The system would automatically generate the YAML file and check it into the VCS, asking the researcher to fill the missing values into this file.
Thereafter, additional checks can be done. In particular, the system checks the file for standards adherence, e.g., tests the numeric values if certain attributes are out of range, tests non-numeric characters, etc. The researcher also receives information about simple statistical (min, max, average and standard deviation) computations. While we have not concretized our technology stack, the usage of continuous integration (CI) runners or Travis has been under consideration for the quality monitoring framework. Our architecture will rely on continuous integration hooks in the VCS server that will be used to drive the workflow for quality checks. This will enable us to develop semantic ontologies to assess the quality of the dataset which can be subjected to data mining techniques in future.
7 Conclusion
In this paper we have described the Conquaire project, whose objective is to develop a generic design infrastructure to support continuous quality control of research data and enable analytical reproducibility of research results. Thus far, we have defined the basic abstract architecture and, to showcase the functionality of the Conquaire system, we have provided a first implementation that outlines the behaviour of the system for a test CSV file.
We are currently working in tandem with research groups at Bielefeld University from diverse fields such as Biology, Chemistry, Psychology, Economics, Sports Science, Computer Science and Robotics. In the near future, we will elicit specific requirements from each of these project partners, set up private repositories for each research project to have a representative sample of different research projects that can guide the development of the Conquaire architecture and system to make sure that it can support more research projects and guide them towards improving the readiness-to-use of their data and also the reproducibility of their analytical workflows. The research artifacts like data, analytical workflows, tools, methods, software, publications, etc., within the Conquaire system will be linked to the trusted data repository PUB, in order to enable the researcher to publicly share and publish their research.
The Conquaire infrastructure described in this paper is a generic design containing a pluggable set of libraries for the framework to facilitate easy adaptation, extension and reuse by other research projects. While the focus is on developing methods and tools to capture and evaluate most of the available data types, we are aware that it is neither practical nor possible to write libraries to support every existing data format. However, the abstract infrastructure architecture facilitates the addition of plugins into the Conquaire infrastructure with minimal effort.
The Conquaire infrastructure software will be under a Free/Libre software license and it is planned to use continuous integration (CI) for building future versions of the Conquaire software itself. However, at the moment we don't know which part of the libraries proposed to be built into the framework will be made persistent when the Conquaire project ends. The design of Conquaire will enable our case study partners to have the freedom to manage and decide the level of migration, maintenance and release of their research data (results, scripts, etc.) generated by their project.
Acknowledgements
This work is part of the Conquaire project, which is funded by the German National Science Foundation (DFG)[8]. We thank the researchers from Bielefeld University who provided insight and expertise that greatly assisted our research. We would like to acknowledge the discussions and feedback received from the project partner members of the Conquaire research project.
Bibliography
| [1] |
Nosek B., et al., Open Science Collaboration: Estimating the reproducibility of
psychological science. Science, 28 August 2015. https://doi.org/10.1126/science.aac4716 |
| [2] |
Florian Prinz, Thomas Schlange & Khusru Asadullah: Believe it or not: How much can we rely on published data on potential drug targets? Nature, September 2011. https://doi.org/10.1038/nrd3439-c1 |
| [3] |
Baker M.: Is there a reproducibility crisis? Nature, 2016. https://doi.org/10.1038/533452a |
| [4] |
Philipp Cimiano, John McCrae, Najko Jahn, Christian Pietsch, Jochen Schirrwagen, Johanna Vompras & Cord Wiljes: CONQUAIRE — Continuous quality control for research data to ensure reproducibility: An institutional approach. Project proposal. https://doi.org/10.5281/zenodo.31298 |
| [5] |
Martin Fenner, Thinking about CSV. |
| [6] |
Greg Wilson, Jennifer Bryan, Karen Cranston, Justin Kitzes, Lex Nederbragt, Tracy Teal, Good Enough Practices in Scientific Computing, ArXiv.org, 2016, arXiv:1609.00037v1 [cs.SE]. |
| [7] |
Greg Wilson: Software Carpentry. Lessons learned. F1000Research, 2016. |
| [8] |
DFG-funded research project number 277747081 (German Abstract). |
About the Authors
Vidya Ayer is a scientific researcher at Bielefeld University, Germany. Her research interest is focused on topics related to knowledge organization, computational reproducibility, computational science, research data management and BigData. She worked as a software engineer professionally and her educational background includes mathematics and technology. She obtained her graduation degree and an Advanced Diploma in Computer Applications from India. Vid volunteers in the free and open source software (FOSS) community and has been an active contributor to the Python, Julia, Ubuntu and Debian projects since 2005.
Christian Pietsch studied linguistics and informatics in Leipzig, Germany, and Edinburgh, Scotland, and holds a master's degree (diplom) in computational linguistics from Saarland University. His professional experience includes two years as a scientific software engineer in the Natural Language Generation group at the Open University in Milton Keynes, England, and four years as a researcher at the Center of Excellence in Cognitive Interaction Technology (CITEC) of Bielefeld University, Germany. Since 2012, Christian has filled various roles at Bielefeld University's department for Library Technology and Knowledge Management where he currently helps researchers improve the replicability of their results, preserves research data and facilitates the introduction of ORCID in Germany.
Johanna Vompras received the graduation degree in computer science (Diplom) from Bielefeld University and the Ph.D. degree from the Heinrich-Heine University in Duesseldorf. Since 2010, she is working at the Bielefeld University Library on services for a university-wide Research Data Management where she is responsible for proving and establishing best practices for the handling of research data, as well as the development of information infrastructures for documentation, archiving, and reuse of research data and their embedding into institutional services.
Jochen Schirrwagen is a research fellow at Bielefeld University Library, Germany. He has a scientific background in Computer Engineering. He worked for the Digital Peer Publishing Initiative for Open Access eJournals at the academic library center "hbz" in Cologne (2004-2008). Since 2008 he is working for DFG and EU funded projects, like DRIVER-II and OpenAIRE. Jochen is interested in the application of metadata information using semantic technologies for the aggregation and contextualization of scientific content.
Cord Wiljes studied Chemistry and Philosophy at Bielefeld University. He worked as a project manager at Bertelsmann AG and subsequently as a freelance IT consultant. In 2011 he joined the Semantic Computing Group at CITEC, where he is working as researcher data manager, developing new approaches for the semantic representation of research data.
Najko Jahn works as Data Analyst at the State and University Library Göttingen, Germany.
Philipp Cimiano received the graduation degree in computer science and computational linguistics from Stuttgart University, Stuttgart, Germany, and the Ph.D. degree from the Karlsruhe Institute of Technology, Karlsruhe, Germany. He is a Full Professor of Computer Science with Bielefeld University, Bielefeld, Germany, and with the Cognitive Interaction Technology Excellence Center. He leads the Semantic Computing Group which performs research in the fields of semantics, ontologies, knowledge acquisition and representation, research data management, as well as cognitive modeling of language acquisition and other cognitive processes. Dr. Cimiano is an Associate Editor of the Journal of Web Semantics, Semantic Web Journal, the Journal of Applied Ontology, and Semantic Computing Journal.