 |
D-Lib Magazine
January/February 2015
Volume 21, Number 1/2
Table of Contents
A Framework Supporting the Shift from Traditional Digital Publications to Enhanced Publications
Alessia Bardi and Paolo Manghi
Istituto di Scienza e Tecnologie dell'Informazione, Consiglio Nazionale delle Ricerche, Italy
{alessia.bardi, paolo.manghi}@isti.cnr.it
DOI: 10.1045/january2015-bardi
Printer-friendly Version
Abstract
Enhanced publications (EPs) can be generally conceived as digital publications "enriched with" or "linking to" related research results, such as research data, workflows, software, and possibly connections among them. Enhanced Publication Information Systems (EPISs) are information systems devised for the management of EPs in specific application domains. Currently, no framework supporting the realization of EPISs is known, and EPIs are typically realized "from scratch" by integrating general-purpose technologies (e.g. relational databases, file stores, triple stores) and Digital Library oriented software (e.g. repositories, cataloguing systems). Such an approach is doomed to entail non-negligible realization and maintenance costs that could be decreased by adopting a more systemic approach. The framework proposed in this work addresses this task by providing EPIS developers with EP management tools that facilitate their efforts by hiding the complexity of the underlying technologies.
1 Introduction
The traditional scholarly communication model is based on the pair document — metadata, where a document is generally a digital scientific article and the relative metadata is provided as a set of structured information supporting interpretation and discovery of the article. It is becoming today evident ([9-11]) how such a model cannot cope with the requirements of modern scholarly communication, which adds to the research chain different kinds of outputs (e.g. software, datasets, workflows) and aims at different usages of such outputs (e.g. research impact measurement, repetition of experiments).
In many disciplines the publication is considered just the tip of the iceberg: the publication describes a research investigation, but it does not provide sufficient information to validate its results. In order for an experiment to be replicable and reproducible, data and processes should be shared as well. In other words, data and processing workflows should be considered "first class citizens", together with publications, of the scholarly communication chain [6, 9, 10, 12, 13, 14, 15, 16, 20, 21]. As a matter of fact, scholarly communication tools should support researchers at sharing any form of research outputs that is relevant to the correct interpretation, re-use (repetition, reproduction), and assessment of research activities [11].
As shown by the survey conducted in [2], Enhanced Publications (EPs) can be generally conceived as digital publications "enriched with" or "linking to" related research results, such as research data, workflows, software, and possibly connections among them. Enhanced Publication Information Systems (EPISs) are systems devised for the management of EPs [3-8, 19]. The majority of those systems are tailored to their specific communities and realized "from scratch" so that functionalities that are shared across disciplines and user communities are re-implemented every time. In fact, EP-oriented software is realized by integrating technologies that are general-purpose (e.g. databases, file stores) and Digital Library-oriented (e.g. repository software, cataloguing systems). The resulting products are often not flexible enough to be adapted to the evolving requirements of the community they target and hardly re-usable and configurable to be re-used in different application domains with similar requirements.
Such a "from scratch" approach entails non-negligible realization and maintenance costs that could be decreased by adopting a more systemic approach, as it had been done in the past with Database Management Systems (DBMSs). Indeed, the success of DBMSs is due to their capability of delivering functionalities that were previously implemented every time by applications dealing with data management issues (e.g. structure, storage, query, optimization, static typing, integrity, validation, redundancy, etc.). The adoption of a DBMS allows database developers to focus their efforts on domain-dependent requirements, rather than on implementing yet another data storage and/or retrieval layer [17][18].
A similar systemic approach has not yet been applied to the EPISs scenario, where ''from scratch" realization is the norm. In this paper we propose the realization of an Enhanced Publication Management System (EPMS) that is a software framework that plays the role of DBMSs in the world of EPs. The framework supports developers of EPISs with tools that (i) hide the complexity of the implementation of domain-independent requirements, (ii) allow the definition of personalized EP data models, (iii) support the realization and configuration of functionalities based on the defined EP data model.
The paper is organized as follows: Section 2 introduces a meta-model for Enhanced Publications, in principle mirroring what the relational data model is for RDMBSs; Section 3 introduces the general requirements of Enhanced Publication Management Systems; Section 4 drafts the architecture of our framework, addressing the requirements defined in Section 3 over the meta-model defined in Section 2.
2 Data models for enhanced publications
In the following we shall refer to the following definition of enhanced publication presented in [2] (in turn a refinement of the definition given by SURF Foundation in [1]:
[Enhanced publications are] digital objects characterized by an identifier (possibly a persistent identifier) and by descriptive metadata information. The constituent components of an EP include one mandatory textual narration part (the description of the research) and a set of interconnected sub-parts. Parts may have or not have an identifier and relative metadata descriptions and are connected by semantic relationships.
The survey conducted in [2] examined several EPISs in order to provide (i) a classification of such systems in terms of the functionalities they offer and (ii) highlight the common features of existing EP data models. The study identified four main classes of EPISs functionalities:
- Packaging of related research assets;
- Web 2.0 reading capabilities;
- Interlinking research outputs;
- Re-production and assessment of scientific experiments.
Moreover, by analyzing the data models adopted by such systems, the investigation characterized EP data models in terms of the following types of enhanced publication parts:
- Embedded parts: identity-less parts that are packaged within the enhanced publication. Embedded parts are not directly discoverable, i.e. it is possible to access them only once the embedding resource has been discovered. Typical examples of embedded parts are the supplementary material of scientific articles managed by journals: you cannot search for supplementary material, but once you have found the relative article (i.e. its embedding part), then you can access it.
- Structured-text parts: (semi-) structured textual documents, such as XMLs.
- Reference parts: remote resources referenced via resolvable identifiers such as URLs or a PIDs (e.g. handle, doi).
- Executable parts: parts that can be executed, such as workflows, web service instances, software code.
- Generated parts: parts that are generated from other parts. Examples are dynamic tables that are updated when the underlying data set changes; or a molecule 3D rendering generated by running a 3D molecule viewer with appropriate parameters.
Based on the above features, we have defined the EP meta-model in Figure 1. The EP meta-model provides the primitives for the definition of custom EP data models (in principle, all EP data models studied in [2] can be expressed as instances of this meta-model), and will inspire the construction of an EP data model definition language of our framework. To draw a parallel with relational databases, the EP meta-model corresponds to the relational model, while the EP data model language corresponds to the SQL definition language.
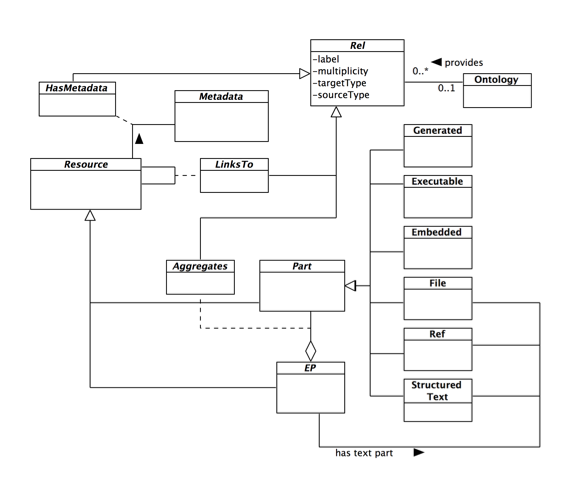
Figure 1: The Enhanced Publication meta-model
EP is a Resource that Aggregates at least one narration (via the has text part relationship) and zero or more other Parts. A narration represents a digital publication to enhance and can be of type File (i.e. a file locally hosted), Ref (i.e. a link to a remote file), or Structured Text (e.g. JATS XML). Parts are Resources that reflect the data model features described above: embedded parts (Embedded), structured-text parts (Structured Text), reference parts (Ref), executable parts (Executable), and generated parts (Generated). Resources can have a unique identifier and descriptive metadata. Resources can be linked with each other via relationships (Rel). The attribute label, which can be free-text or a term from an Ontology, expresses the semantics of a relationship.
In the following, two examples are given to show how EPIS developers can represent an EP data model in terms of the meta-model.
Example 1
Let's suppose we want to define a model for Enhanced Publications that allows a PDF publication to be connected to the workflows used to generate research data. Figure 2 shows a possible implementation where research data are deposited at a remote location (e.g. a thematic data repository). Referenced research data is connected to its generating workflow via the relationship class LinksTo. In this case the semantics of the relationship has been plugged in as simple free text (the label generatedBy).
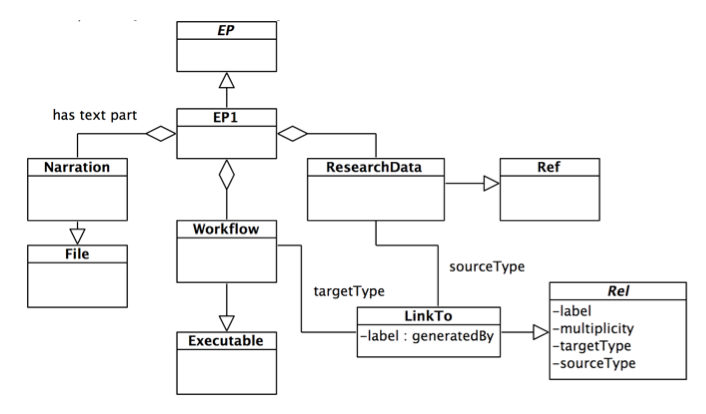
Figure 2: Example 1: a data model for EPs composed of remote research data and executable workflows
Example 2
Let's suppose we want to extend example 1 to:
- Allow different types of research data: we would like the model to allow data to be local or remote files (e.g. spreadsheets) or entries from a remote database (e.g. a protein in UniProtKB).
- Include the CERIF-compliant ontology FRAPO (Funding, Research Administration and Projects Ontology) to define the semantics of the association between workflows and research data.
A possible solution shown in Figure 3: research data is modeled as a Resource that aggregates local and remote research data files and database queries, while the relationship class LinksTo is defined to be provided by the FRAPO ontologies, so as to narrow down the allowed labels to be terms from the FRAPO ontology.
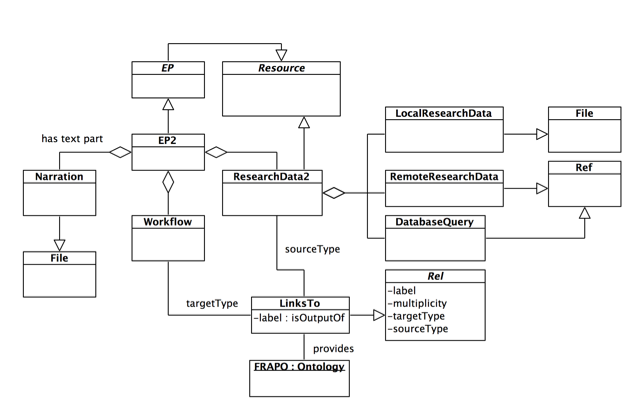
Figure 3: Example 2: a data model for EPs where research data can be local files, remote files and entries from databases
3 Requirements of Enhanced Publication Management Systems
In addition to the primitives for the definition of EP data models, an EPMS should also provide functionality for EP management that hides the complexities of underlying technologies. Some functions are very generic, as they apply to any data management system, while others are more specific to the scholarly communication world and the EP scenario.
General requirements
Grad and Bergin provide in [17] a summary of the functionality that made DBMSs so popular:
- They hide the complexity needed to store and retrieve data on different back-ends.
- They provide data access and retrieval functions ready to use by different applications.
- They enable data sharing among different applications and users.
- They offer user-oriented languages for data definition, manipulation, and access.
- They support data and application portability.
Those functionalities must also be provided by an EPMS in order to support developers with tools for the setup, operation and maintenance of EPISs.
Requirement 1. Supporting different back-ends for data storage.
The framework should provide a storage management module for the configuration of the storage back-ends to use. Depending on the functional requirements of the target EPIS, a type of back-end may be preferable to another. For example, a triple store could be more suitable to support the export of EPs via Linked Data, while a full-text index could be dedicated to support search on the text and on the metadata of publications.
Requirement 2. Offering data definition, manipulation, and access languages.
The framework should provide a language for the definition of EP data models (EP-DMDL, EP Data Model Definition Language). In addition, developers of EPISs should be able to operate on EP instances (compliant to the defined EP data model) with a dedicated domain-specific language that allows manipulation of resources whose types are defined in the EP data model (EP-DSML, EP Domain Specific Manipulation Language).
Requirement 3. Enabling data sharing.
The framework should support the export of content via different standard APIs and protocols to serve third-party applications. Examples are OAI-PMH, OAI-ORE, Linked Data, OpenSearch, RDF.
Requirement 4. Supporting data portability.
To further promote data sharing and re-use, the framework should support open standards for the representation of data. A data transformation module may take care of transforming data from one format to another, ensuring a high level of interoperability and portability.
EP-specific requirements
Requirement 5. Support the integration of heterogeneous data sources.
EPs are digital objects that aggregate research outputs and related material. Such content is already available from different types of data sources, such as literature and data repositories, archives, metadata aggregators, and scientific databases. Data sources export different typologies of content according to different formats and via different protocols. EPMSs should support developers in the integration of such diverse content, providing (i) built-in functionalities for the import of content via standard protocols and (ii) supporting the implementation of ad-hoc integration functions, for those data sources exporting content via idiosyncratic protocols and formats.
Requirement 6. Support the management of dynamic data sources.
Data sources are typically dynamic in terms of (i) supported protocols and formats and (ii) availability of services and content. Furthermore, the list of integrated data sources may change during the lifetime of an EPIS. EPMSs should therefore provide data source management functionality to ease the administrative operations needed to take care of the dynamic nature of the data sources.
Requirement 7. Support the integration of content.
Due to the heterogeneity of the content, a transformation and harmonization module is necessary in order to massage the incoming material and transform it in a homogeneous format, so that further operations can be performed on content without tackling again the peculiarities of each data source.
Requirement 8. Enable the customization of the EP data model.
Components of EPs in one application domain may be very different from the components of EPs in another domain, both in terms of structure and semantics. Therefore, an EPMS should not provide a pre-defined model, as it would be either too generic or too specific. A generic EP data model would be usable in any domains, but it would not cope with specific requirements of the target community. On the other hand, if the model captures some community-specific requirements, it would be not usable in different contexts. EPMSs should instead offer tools for the definition of EP data models, so that EPIS developers can define the structure and semantics of the EPs they want to manage.
Requirement 9. Support the enrichment and curation of content.
When we create EPs, we are not just creating a new digital object by composing other digital objects. We are also creating new relationships between objects that can be useful to better the quality of the EPs and enrich the original content. For example: let's suppose we have imported metadata records about the same scientific article from two different data sources. If we can understand that they are indeed about the same article, then it is possible to eliminate duplicates by merging the two records into one, so as to better the quality of the integrated content.
4 The framework
The framework has been designed to satisfy both the general and EP-specific requirements. The framework is organised in five functional areas as depicted in Figure 4:
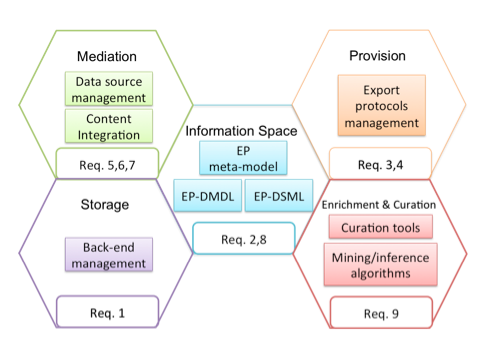
Figure 4: Functional areas of the framework
Mediation area: research materials to use for the creation of enhanced publications are already available from different data sources, such as existing digital libraries, institutional and data repositories. Different data sources typically offer heterogeneous export interfaces in terms of:
- Exchange protocols (e.g. JDBC, OAI- PMH, Web Services);
- Format of data (e.g. PDF, XML, CSV) and metadata (e.g. JSON, XML);
- Data model (ranging from standards such as Dublin Core, EAD, and CERIF, to idiosyncratic data models).
The mediation area copes with the heterogeneity and dynamicity of the data sources and supports the realization of functions to integrate content into the EPIS. Moreover, the mediation area acts as proxy for the content that cannot be directly imported. This is the case for large scientific database and big data warehouses.
Storage Area: EPISs might want to integrate different back-end storage technologies for different purposes. For example, a triple store could be more suitable to support the export of EPs via Linked Data, while a full-text index could be dedicated to support search on the text and on the metadata of publications. An EPMS should be able to hide the complexity of the underlying technologies but, at the same time, EPIS developers should be allowed to tune the back-end storage configuration, when needed. For example, EPIS developers may want to decide to add an index to a relational database, or to configure how many replicas the EPIS must keep to ensure that the required level of redundancy is reached.
Enrichment & Curation Area: Enhanced Publications can be enriched and curated in order to increase data quality. The enrichment and curation area includes all functional elements at this purpose, for example: text mining algorithms for knowledge extraction from digital publications (e.g. for mining links to referenced datasets or to taxonomy entries), modules for calculating statistics about data quality, algorithm to detect and merge duplicate records.
Provision Area: includes functional elements for the export of Enhanced Publications. Provision modules that implement different exchange protocols (e.g. OAI-PMH, OAI-ORE, OpenSearch) and deliver EPs in different formats can be activated to serve heterogeneous consumers. For example, an OAI-ORE provider can be configured to export EPs as aggregations of digital publications and supplementary material; HTML5 files may be delivered to a graphical user interface to provide Web 2.0 reading capabilities.
Information Space Area: offers languages for the definition of EP data models (EP-DMDL) and manipulation of EPs compliant to the defined EP data model (EP-DSML). The EP Data Model Definition Language the framework provides is an object-oriented language implemented in Java/Groovy that has been derived from the EP meta-model in Figure 1. Classes of the EP meta-model correspond to Java classes that can be instantiated to define the structure and semantics of the types of resources to include in the EP data model. From the defined EP data model, a domain-specific language for the manipulation of EP instances is derived.
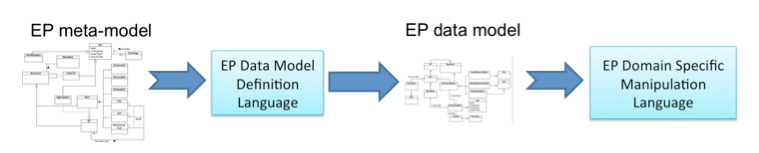
Figure 5: Data model definition language and domain specific manipulation language
In the following, the two EP data models presented in Section 2 are implemented in EP-DMDL.
Example 1
The following EP-DMDL snippet defines the types that form enhanced publications of type EP1: Narration, Workflow, and ResearchData.
EP EP1 = new EP();
File Narration = new File(mime:"application/pdf");
Executable Workflow = new Executable();
Ref ResearchData = new Ref();
EP1.hasTextPart(targetType:Narration);
Aggregates aggregatesWf = new Aggregates(sourceType:EP1,
targetType:Workflow,
label:"generates data via", multiplicity:0..*);
Aggregates aggregatesData = new Aggregates(sourceType:EP1,
targetType:ResearchData, label:"has data", multiplicity:0..*);
LinksTo generatedBy = new LinksTo(sourceType:ResearchData,
targetType:Workflow, label:"generatedBy", multiplicity:0..*);
Once the "skeleton" is ready, the developer can customize the properties of each type by specifying the list of interesting properties inline or by providing an XML schema. For example, the following specification customizes the properties for EP1 with the Dublin Core metadata format. Properties for Workflow are instead defined as an extension of Dublin Core with some idiosyncratic properties.
Metadata dcMetadata = Metadata.fromXSD("http://dublincore.org/schemas/xmls/simpledc20021212.xsd");
Metadata wfMetadata = Metadata.merge(dcMetadata , {
{name:executionTime , type:long , repeatable:0, required:0} ,
{name:executedBy, type:string, repeatable:0, required:1} } );
HasMetadata EPHasMetadata = new HasMetadata(sourceType:EP1, targetType:dcMetadata, label:"descriptiveMetadata");
HasMetadata WfHasMetadata = new HasMetadata(sourceType:Workflow,
targetType:wfMetadata, label:"descriptiveMetadata");
Example 2
The code snippet below shows the definition of the ResearchData2 class and how to select relationships' labels from existing ontologies. Specifically, the term isOutputOf is selected from the CERIF-compliant ontology FRAPO (Funding, Research Administration and Projects Ontology).
Resource ResearchData2 = new Resource();
File LocalResearchData = new File();
Ref RemoteResearchData = new Ref();
Ref DatabaseQuery = new Ref();
LinksTo aggregatesLocal = new LinksTo(sourceType:ResearchData2,
targetType:LocalResearchData, label:"aggregates",
multiplicity:0..*);
LinksTo aggregatesRemote = new LinksTo(sourceType:ResearchData2,
targetType:RemoteResearchData, label:"aggregates",
multiplicity:0..*);
LinksTo aggregatesQuery = new LinksTo(sourceType:ResearchData2,
targetType:DatabaseQuery, label:"aggregates",
multiplicity:0..*);
Ontology FRAPO = Ontology.load("http://purl.org/cerif/frapo");
LinksTo aggregatesQuery = new LinksTo(sourceType:ResearchData2,
targetType:Workflow, label:FRAPO.isOutputOf, multiplicity:0..*);
5 Conclusion
Enhanced Publication Information Systems (EPISs) are typically realized "from scratch" by integrating existing technologies. This approach entails non-negligible realization and maintenance costs. Consequently, the main agents in the realization of running EPISs are those that have human and economic resources to afford such an investment. Examples are publishers such as PLoS, Elsevier, and Nature. Research institutions offering traditional Digital Library Systems to researchers cannot generally afford the realization and maintenance cost of a new information system and thus they are currently excluded from the shift from traditional digital publications to enhanced publications.
We have argued that a more systemic approach would decrease the cost of realization and maintenance of EPISs and support developers with EP management tools that hide the complexity of the underlying technologies.
We have presented the notion of Enhanced Publication Management Systems (EPMSs), frameworks devised to support the realization of EPISs, whose role can be compared to the role of RDBMS in the relational database scenario.
A set of requirements for EPMSs has been listed and a framework that satisfies them has been presented. The framework supports developers of EPISs with tools that (i) hide the complexity of the implementation of domain-independent requirements, (ii) allow the definition of personalized EP data models via an object-oriented language that has been derived from the EP meta-model, i.e. a data model for EP data models, realized based on the results of the survey conducted in [2], (iii) support the realization and configuration of functionalities based on the defined EP data model.
Acknowledgements
This work is partially funded by the European Commission FP7 P project OpenAIREplus (Grant agreement: 283595, Call: FP7INFRASTRUCTURES-2011-2).
References
[1] Woutersen-Windhouwer Brandsma, R., Verhaar, P. , Hogenaar, A., Hoogerwerf, M., Doorenbosch, P., Drr, E., Ludwig, J., Schmidt, B., Sierman, B.: Enhanced Publications: Linking Publications and Research Data in Digital Repositories. SURF / E-Driver (2009).
[2] Bardi, A., Manghi, P.: Enhanced publications: Data models and information systems. LIBER Quarterly 23(4) (2014).
[3] Hoogerwerf, M.: Durable Enhanced Publications. In: Proceedings of African Digital Scholarship & Curation 2009. (2009).
[4] Shotton, D., Portwin, K., Klyne, G., Miles, A.: Adventures in semantic publishing: Exemplar semantic enhancements of a research article. PLoS. http://doi.org/10.1371/journal.pcbi.1000361
[5] Attwood, T., Kell, D., McDermott, P., Marsh, J., Pettifer, S., Thorne, D.: Utopia documents: linking scholarly literature with research data. Bioinformatics (Oxford, England) 26(18) (09 2010) i568—74. http://doi.org/10.1093/bioinformatics/btq383
[6] Schutter, E.: Data publishing and scientific journals: The future of the scientific paper in a world of shared data. Neuroinformatics 8 (2010) 151—153. http://doi.org/10.1007/s12021-010-9084-8
[7] Breure, L., Voorbij, H., Hoogerwerf, M.: Rich internet publications: Show what you tell. Journal of Digital Information 12(1) (2011).
[8] Garcia Castro, L., McLaughlin, C., Garcia, A.: Biotea: Rdfizing pubmed central in support for the paper as an interface to the web of data. Journal of Biomedical Semantics 4(Suppl 1) (2013). http://doi.org/10.1186/2041-1480-4-S1-S5
[9] Gray, J.: Jim gray on e-science: a transformed scientific method. The fourth paradigm: Data-intensive scientific discovery (2009) 19—31
[10] Borgman, C.L.: The conundrum of sharing research data. Journal of the American Society for Information Science and Technology 63(6) (2012) 1059—1078. http://doi.org/10.1002/asi.22634
[11] Castelli, D., Manghi, P., Thanos, C.: A vision towards scientific communication infrastructures. International Journal on Digital Libraries (2013) 1—15. http://doi.org/10.1007/s00799-013-0106-7
[12] King, G.: An introduction to the dataverse network as an infrastructure for data sharing. Sociological Methods & Research 36(2) (2007) 173—199. http://doi.org/10.1177/0049124107306660
[13] Brase, J.: Datacite-a global registration agency for research data. In: Cooperation and Promotion of Information Resources in Science and Technology, 2009. COINFO'09. Fourth International Conference on, IEEE (2009) 257—261. http://doi.org/10.1109/COINFO.2009.66
[14] Ball, A., Duke, M.: How to cite datasets and link to publications. Dcc how-to guides, Edinburgh: Digital Curation Centre (2012).
[15] Tenopir, C., Allard, S., Douglass, K., Aydinoglu, A.U., Wu, L., Read, E., Manoff, M., Frame, M.: Data sharing by scientists: Practices and perceptions. PLoS ONE 6 (6/2011 2011) e21101. http://doi.org/10.1371/journal.pone.0021101
[16] Huang, X., Ding, X., Lee, C.P., Lu, T., Gu, N.: Meanings and boundaries of scientific software sharing. In: Proceedings of the 2013 conference on Computer supported cooperative work, ACM (2013) 423—434. http://doi.org/10.1145/2441776.2441825
[17] Burton Grad and Thomas J. Bergin. Guest editors' introduction: History of database manage- ment systems. IEEE Annals of the History of Computing, 31(4):3—5, 2009. http://doi.org/10.1109/MAHC.2009.99
[18] T. Haigh. How data got its base: Information storage software in the 1950s and 1960s. Annals of the History of Computing, IEEE, 31(4):6—25, Oct 2009. http://doi.org/10.1109/MAHC.2009.123
[19] Sean Bechhofer, David De Roure, Matthew Gamble, Carole Goble, and Iain Buchan. Research objects: Towards exchange and reuse of digital knowledge. The Future of the Web for Collaborative Science, 2010. http://doi.org/10.1038/npre.2010.4626.1
[20] Stodden, Victoria, Friedrich Leisch, and Roger D. Peng, eds. Implementing Reproducible Research. CRC Press, 2014.
[21] Stodden, Victoria, & Sheila Miguez. "Best Practices for Computational Science: Software Infrastructure and Environments for Reproducible and Extensible Research." Journal of Open Research Software [Online], 2.1 (2014): e21. http://dx.doi.org/10.2139/ssrn.2322276
About the Authors
 |
Alessia Bardi received her MSc in Information Technologies in the year 2009 at the University of Pisa, Italy. She is a PhD student in Information Engineering at the Engineering Ph.D. School "Leonardo da Vinci" of the University of Pisa and works as graduate fellow at the Istituto di Scienza e Tecnologie dell'Informazione, Consiglio Nazionale delle Ricerche, Pisa, Italy. Today she is a member of the InfraScience research group, part of the Multimedia Networked Information System Laboratory (NeMIS). She is involved in EC funded projects for the realisation of aggregative data infrastructures. Her research interests include service-oriented architectures and data infrastructures for e-science and scholarly communication.
|
 |
Paolo Manghi is a researcher at Istituto di Scienza e Tecnologie dell'Informazione, Consiglio Nazionale delle Ricerche, Pisa, Italy. He received his PhD in Computer Science at the University of Pisa (2001). Today he is a member of the InfraScience research group, part of the Multimedia Networked Information System Laboratory (NeMIS). His current research interests include data ICT infrastructures for science and technologies supporting modern scholarly communication. He is technical manager of the OpenAIRE infrastructure (www.openaire.eu).
|
|